|
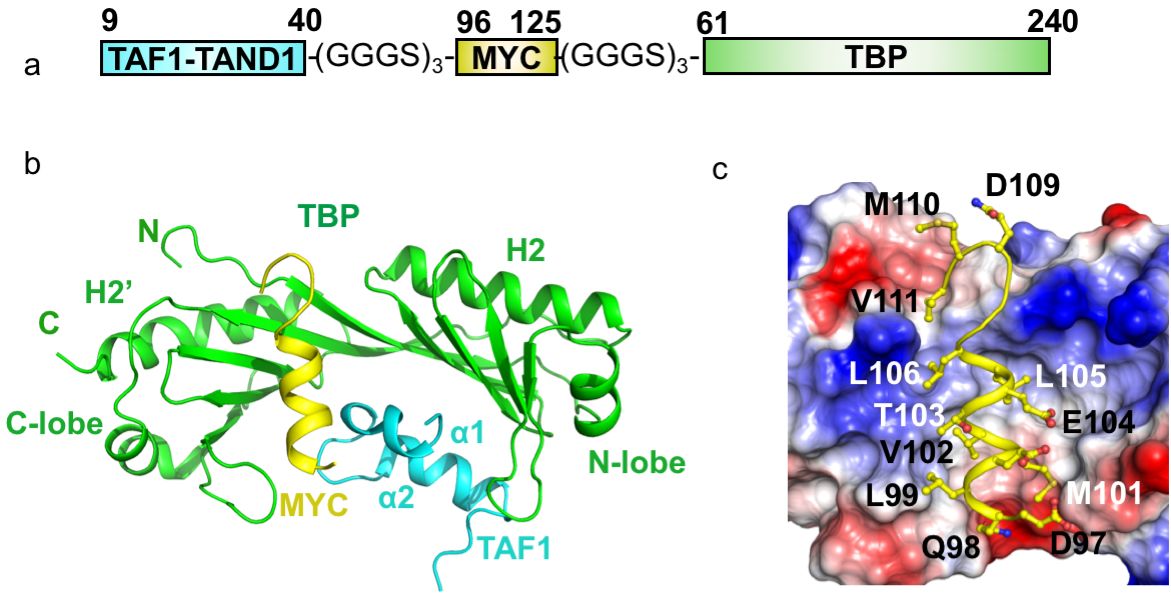
In our recent Nature Structural & Molecular Biology paper (Nov. 2019) on the interaction of the Myc oncoprotein with TATA-box binding transcription factor TBP, in collaboration with the Penn (Toronto, Canada) and Sunnerhagen (Linköping, Sweden) labs, we used a fusion protein approach to make a tripartite fusion protein that finally yielded a complex structure, which formed the cornerstone of the discovery described in the paper.
We used three copies of the GGGS motif for each linker between the target proteins. These linkers are so flexible that they did not show up in the electron density map and shall not impose any structural restraints. Several lines of evidence suggested the observed crystal structure of the fusion protein is physiologically relevant and not an artifact:
The reason we adopted this fusion protein approach is multi-faceted:
The fusion protein approach is versatile and has been used previously in other works, for example, in the structure of an Arf6 GTPase and ArfGAP(ASAP3) complex, where the affinity of the complex is low. However, this approach must be combined with other validation methods to confirm the interfaces observed in the structure are physiologically relevant. One such case of an artifact caused by the fusion approach is our unpublished structure (released in the public domain) of the fusion protein of Arf1 and ArfGAP1 (PDB:3O47), in which the interface is neither consistent with that of the Arf6/ASAP3 complex nor with an early report of the complex structure on Cell (1999). The choice of linker is also worth considering. In the PDB:3O47 structure, we used a natural intrinsically disordered peptide (IDP) region of ArfGAP1 to serve as the linker to obtain the structure.
0 Comments
Plagiarism, including self-plagiarism, is a serious offence in academics. UWindsor Leddy library has a list of tips on avoiding plagiarism.
SafeAssign is a plagiarism checking service available for UWindsor. However, it is slightly tricky to access the service. It is available in the Blackboard Learn system. Details can be found at the Wiki-page of UWindsor Blackboard Learn. "Direct submit" in the BB Sandbox course, is a quick way to check one's writing to avoid unintended plagiarism. It is not necessary to be a course assignment to use this service. |
Twitter
|
 RSS Feed
RSS Feed
